Institute of Evolution, University of Haifa, Haifa 31905, Israel Contributed by Eviatar Nevo, November 9, 2005
The Dead Sea is one of the most saline lakes on earth (≈340 g/liter
salinity) and is ≈10 times saltier than the oceans. Eurotium herbariorum, a common fungal species, was isolated from its water. EhHOG gene, encoding a mitogen-activated protein kinase (MAPK) that plays an essential role in the osmoregulatory pathway in yeast and many other eukaryotes, was isolated from E. herbarium. The deduced amino acid sequences of EhHOG indicated high similarity with homologous genes from Aspergillus nidulans, Saccharomyces cerevisiae, and Schizosaccharomyces pombe and contained a TGY motif for phosphorylation by MAPK kinase. When EhHOG was expressed in S. cerevisiae hog1Δ mutant, the growth and aberrant morphology of the hog1Δ mutant was restored under high osmotic stress conditions. Moreover, intracellular glycerol content in the transformant increased to a much higher level than that in the mutant during salt-stress situations. hog1Δ mutant complemented by EhHOG outperformed the wild type or had higher genetic fitness under high Li+ and freezing-thawing conditions. The present study revealed the putative presence of a high-osmolarity glycerol response (HOG) pathway in E. herbarium and the significance of EhHOG in osmotic regulation, heat stress, freeze stress, and oxidative stress. The Dead Sea is becoming increasingly more saline while the fungi living in it evolutionarily adapt to its high-saline environment, particularly with the extraordinarily high Li+ concentration. The Dead Sea is potentially an excellent model for studies of evolution under extreme environments and is an important gene pool for future agricultural genetic engineering prospects.
Eurotium herbariorum | HOG | salinity | genetic resources | crop improvement
| T |
The Dead Sea is one of the most saline lakes on earth (salinity ≈ 340 g/liter). The studies on the biology of the Dead Sea revealed a variety of microorganisms including red halophilic Archaea, unicellular green algae (Dunaliella parva Lerche), different types of bacteria, and possibly even protozoa (1). Recently, filamentous fungi were isolated from surface water to 300 m down in the Dead Sea (2). Although the isolated fungi did not grow in undiluted Dead Sea samples, they showed a remarkable salt tolerance and, in many cases, even had a requirement for high salt concentrations, making them halophilic. Eurotium herbarium is the most common species isolated from the Dead Sea water, from the surface to a depth of 300 m in all investigated seasons (3). All of these organisms need to adapt to the extremely high salinity of the Dead Sea brines. Exposure to high environmental osmolarity leads to dehydrate-creases. To cope with this challenge, the cells of both prokaryotic and eukaryotic microorganisms have developed mechanisms to adapt to severe osmotic changes in their environments, often called osmoregulation. To adapt to salt stress, microorganisms balance high external osmotic pressure by synthesizing and/or accumulating low-molecular-mass compounds, which are compatible with cellular function and do not inhibit enzymes (4). Increased synthesis and/or accumulation of glycerol and other compatible solutes, mainly polyols, have been the major features of fungi osmoregulation (5). Mitogen-activated protein kinase (MAPK) is a key component of the evolutionarily conserved signal transduction cascades consisting of MAPK/extracellular signal-related kinase (ERK) activated by a MAPK/ERK kinase (MEK), which in turn is activated by a MEK kinase (6). Eukaryotic organisms use different MAPK cascades to regulate various aspects of cellular function (7, 8). MAPKs that specifically transmit environmental stress signals are also known as stress-activated protein kinases (SAPKs). This pathway is called the high-osmolarity glycerol (HOG) response pathway in Saccharomyces cerevisiae (9). Members of this MAPK subfamily include Hog1 in S. cerevisiae, Spc1 (also called StyI) in Schizosaccharomyces pombe, SakA in Aspergillus nidulans, and p38/JNK in mammals. Indeed, S. cerevisiae hog1 mutants are sensitive to high osmolarity, whereas spc1 mutations in S. pombe result in sensitivity to high osmolarity, heat shock, and oxidative stress. Activation of the HOG pathway increases the transcription of some proteins, including enzymes involved in glycerol synthesis (10). As a result, a high accumulation of glycerol inside the cell occurs and leads to increased internal osmolarity and restores the osmotic gradient between the cells and their environment (11). Therefore, the HOG1 gene holds a key position in co-adaptation of the yeast
S. cerevisiae.
The presence of HOG1 homologous genes has been reported in fungal species (12, 13) and mammals (14), indicating that this pathway is conserved among eukaryotes. However, no information is available for the HOG pathway or the molecular mechanism of stress tolerance in E. herbarium. Here, we cloned the gene EhHOG of the filamentous fungus E. herbarium from the Dead Sea and then transformed it into a wild-type (WT) and hog1 mutant of the baker’s yeast S. cerevisiae. The expression of EhHOG functionally complemented S. cerevisiae hog1 mutant, and, remarkably, the complemented hog1 mutant displayed higher tolerance to several stresses than that of the WT strain of
S. cerevisiae. Consequently, we propose the genetic resource of the Dead Sea fungi as a potentially promising resource to improve salt tolerance in other organisms in relation to various biological systems, including saline agriculture.
Materials and Methods
Fungus, Yeast Strains, and Culture Conditions. E. herbariorum, isolated from the Dead Sea water, was cultured in 1% yeast extract/10% glucose medium (GY) at 25°C (3). Escherichia coli
Conflict of interest statement: Carmel-Haifa University Economic Corp. Ltd. has filed for a patent on EhHog in the names of Y.J., S.W., and E.N.
Abbreviations: ERK, extracellular signal-related kinase; HOG, high-osmolarity glycerol; MAPK, mitogen-activated protein kinase; SAPK, stress-activated protein kinase.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ229154).
*To whom correspondence should be addressed. E-mail: nevo@research.haifa.ac.il.
© 2005 by The National Academy of Sciences of the USA strain XL/B was used for transformation and plasmid propagation. E. coli XL/B was grown in Luria–Bertani (LB) medium at 37°C for plasmid manipulation. The S. cerevisiae strains used in this study were WT YSH6.142–3A (MATa leu2–3/112 ura3–1 trp1–1 his3–11/15 ade2–1 can1–100 GAL SUC2 mal0) and YSH444 (MATa hog1Δ::TRP1 mutant) (10). Yeast cells were grown in a rich medium containing 1% yeast extract, 2% peptone, and 2% dextrose (YPD) or in a minimal medium containing 0.67% yeast nitrogen base and 2% (wt/vol) glucose (SD) supplemented with required amino acids on a rotary shaker at 30°C (15). Agar (1.5%) was added to these media when required.
DNA and RNA Analysis. Mycelia of E. herbarium cultured in GY medium for 2 weeks and harvested by filtration were then soaked in fresh GY medium with 2 M NaCl for various periods. Total RNA was prepared by using the Total RNA Isolation Kit (Beit Haemek, Ashrat, Israel). DNA from E. herbarium was isolated and purified by modifying the Raeder and Broda method (16). The genomic DNA of yeast was isolated according to basic protocol (17). For Southern blot analysis, 10 µg of high-purity genomic DNA was digested with enzymes EcoRI, HindIII, and BamHI (New England Biolabs) and then transferred to a Hybond N nylon membrane (Amersham Pharmacia). Southern hybridizations (60°C) were carried out as described by Sambrook et al. (18). For the probe, an EhHOG-containing DNA fragment (≈800 bp) was obtained by PCR amplification with oligonucleotides 5′-AAGAAGATTATGAAGCCTT- TCAGC-3′ and 5′-CATAATTTTCCATGTGTCGACCGG-3′
as the primers and was labeled with [32P]dCTP by a random primer-labeling kit (Beit Haemek). For Northern blot analysis, the method of hybridization (68°C) was the same as for Southern blots.
Isolation of EhHOG Gene from E. herbarium. To isolate the HOG gene from E. herbarium, PCR was performed by using genomic DNA as the template and sequences 5′-ATGGCGGAAT- TCGTGCGTGCCACGATT-3′ and 5′-GGCCGCGAATGC-
CTGCTGGCCATCCCC-3′ as the primers for 35 cycles each consisting of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C. The amplified fragments were subcloned into pGEM-T easy (Pro- mega) and sequenced. To obtain full-length HOG cDNA, RT- PCR was performed. For RT-PCR, two HOG-specific primers (5′-CAAAGCTTATGGCGGAATTCGTGCGTGCCAC- GATT-3′ and 5′-GGCCGCGAATGCCTGCTGGCCATC-
CCC-3′) were synthesized. First-strand cDNA synthesis was performed in a 20-µl reaction mixture according to the cDNA Synthesis system (Promega). A 1-µl sample of first-strand cDNA products was then used as template DNA in PCR for E. herbarium HOG cDNA. PCR amplification was performed with 50-µl reaction that contained 5 µl of 10× buffer, 0.2 mM dNTPs (2 µl of 5 mM dNTPs), 1 µl each of 10 µM HOG-specific primer, and 2 units of Taq polymerase (Biotools, Madrid) for 30 cycles each consisting of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C. The amplified fragments were subcloned into pGEM-T Easy and sequenced.
Phylogenetic Analysis. Phylogenetic trees based on the amino acid sequence alignment of 36 MAPKs were constructed by using the neighbor-joining method (19) implemented in MAGA2.1 (20). The reliability of the tree topology was evaluated by bootstrap analysis of 1,000 replicates. The resulting estimated amino acid distances were corrected for multiple amino acid substitutions per single site by Poisson correction.
Function Expression of EhHOG in S. cerevisiae. For expression of EhHOG in S. cerevisiae, 1.1-kb full-length EhHOG cDNA was inserted into HindIII/NotI sites under transcriptional control of the promoter for alcohol dehydrogenase (ADH) in plasmid pADNS resulting in plasmid pADNS–EhHOG. Plasmid pADNS harboring the S. cerevisiae HOG1 coding sequences was obtained by subcloning HindIII–NotI fragments, which were obtained by amplification from S. cerevisiae genomic DNA through primers 5′-ACAAAGCTTATGACCACTAACGAGGAATT-3′ and
5′-CTGGCGGCCGCTTACTGTTGGAACTCATTAG-3′.
Transformation of yeast cells with pADNS–EhHOG was performed by the lithium acetate/single-stranded carrier DNA/ poly(ethylene glycol) methods (21).
Growth Assays. The EhHOG gene was expressed in S. cerevisiae WT strain YSH6.142–3A and mutant YSH444 (MATa hog1Δ::TRP1), which lacks HOG1. Cultures of strains trans- formed with pADNS or pADNS-EhHOG were grown in SD- LEU medium and YPD medium in the presence or absence of various concentrations of NaCl, LiCl, and sorbitol. For the growth test, cells were grown at 30°C for 36 h; serial dilution, 1/10, was made at each step. From each dilution, 10 µl was spotted on different media and incubated at 30°C for 3–5 days. At various time intervals, aliquots of the cultures were measured by optical density at 600 nm. For heat stress, cells were grown to OD600 1.8 –2. Serial dilutions of 10 times were made. Ten microliters of each dilution were spotted on solid YPD plates. Cells on YPD plates were incubated at 42°C for 1 day and allowed to recover at 28°C for 36 and 69 h; for 2-day heat stress, cells were recovered at 28°C for 48 h. Freeze-stress tolerance of cells was determined by measuring viability after the number of freeze-thaw cycles (—30°C and room temperature). Percent survival was expressed relative to the initial viability before freezing. For oxidative stress, cells were grown to OD600 1.0 –1.5 in SD medium at 30°C and then incubated for 1.5 h with increasing concentrations of H2O2 in SD. Percent survival was expressed relative to the initial viability before oxidative stress. Two independent transformants were included, and three independent experiments were performed.
Assays of Intercellular Glycerol Content in Various S. cerevisiae Strains. Cells were cultured at 30°C in SD media and harvested at the early exponential phase (OD600 = 0.5– 0.8). Subsequently, cells were resuspended in a new media with or without 0.4 M NaCl. After incubation for 1 h at 30°C, cells were harvested and prepared as described by Andre et al. (22). The glycerol content was determined according to the application manual of the glycerol-F kit (Roche Molecular Biochemicals).
Results and Discussion
Isolation and Characterization of the EhHOG from E. herbariorum. Successful amplification of HOG1 homolog in E. herbarium was achieved with primers derived from conserved regions of HOG1 from various fungal species available, with flanking regions obtained by chromosome walking (23). Sequence analysis revealed a long ORF interrupted by eight introns, whose positions were confirmed by sequencing cDNA and genomic DNA clones (see Fig. 8, which is published as supporting information on the PNAS website). This ORF predicted 366- amino acid residues with protein masses of 41.62 kDa, which were highly similar to kinase from the stress-activated MAPK family. Therefore, the corresponding gene was named EhHOG (for E. herbarium HOG). The EhHOG protein shows the highest identity to SakA (81% identity, 86% similarity) from A. nidulans and also shares high homology to other SAPKs such as Spc1 (78% identity, 87% similarity) in S. pombe, HOG1 (66% identity, 72% similarity) in S. cerevisiae, and Mpkc (60% identity, 74% similarity) in A. nidulans (Fig. 1). A TGY motif, characteristic of hyperosmolarity-activated MAPKs (24), was found at amino acids 171–173 (Fig. 1). Similar motifs have been found in other yeast species (13) and also in MAPKs from higher

Fig. 1. The EhHOG gene encodes a member of the stress MAPK family. Multiple alignment (CLUSTAL W software, www.ebi.ac.uk/clustalw) of homology-gous proteins from E. herbariorum EhHOG, A. nidulans SakA (AF282891), A. nidulans Mpkc (AF195773), S. pombe STY1 (X89262), and S. cerevisiae Hog1 (U53878). The conserved TGY phosphorylation lip found in stress MAPKs is shaded. Boxed amino acid represents the C-terminal common docking (CD) motif, in which the conserved hydrophobic amino acids tyrosine (Y) and histidine (H) are underlined, and conserved acidic aspartic acids (D) are bold. ✽, perfectly conserved residues; :, very similar residues; •, similar residues.
eukaryotes, which have been shown to complement S. cerevisiae hog1Δ mutants (14, 25). This TGY motif is indicated to be phosphorylated by MAPK kinase (pbs1p) in S. cerevisiae (26). EhHogp is thus likely to be phosphorylated by a kinase similar to pbs2p present in the E. herbarium genome.
A conserved common docking (CD) domain motif of the MAPK family is present in EhHogp (Fig. 1). The CD domain contains two acidic amino acids (Asp-304 and Asp-307) (Fig. 1), which is crucial for docking to a cluster of basic amino acids commonly present in MAPK-docking sites (27). Asp-304, and Asp- 307 of EhHogp could serve to establish critical electrostatic interactions with the positively charged amino acids of docking domains of upstream and downstream effectors together with the amino acids Tyr-302 and His-303. This docking motif in MAPKs is commonly used for recognition of their activators, regulators, and substrates. The docking interaction increases the efficiency of all enzymatic reactions and may help to regulate the specificity of molecular recognition (28).
Phylogenetic analysis using representative amino acid sequences from the three major MAPK subgroups indicated that the sequences were clustered primarily by the type of MAPK, forming three major clades: SAPK, ERK, and MAPK3. Within clades ERK and SAPK, several subgroups were clearly identified. On this tree, EhHOG was tightly clustered with HOG subgroup of fungi relatives (Aspergillus and yeast) (Fig. 2). HOG subgroup is characterized by the TGY motif (Fig. 2), which is found in the sequences of EhHOG (Fig. 1). HOG subgroup appears more specific in their activation by osmotic stress compared with other signals (29). With respect to the high-salinity environment of the Dead Sea, EhHOG may have similar specificity to other HOG genes.
Southern blot analysis of the genomic DNA was performed to estimate the number of EhHOG copies in E. herbariorum. The banding pattern obtained for genomic DNA cut with EcoRI, HindIII, and BamHI suggested the existence of only one copy of the EhHOG gene in the genome (see Fig. 9, which is published as supporting information on the PNAS web site).
Complementation of a S. cerevisiae hog1Δ Mutant by EhHOG in High-Salt Stress. To determine the functions of EhHOG, comple- mentation of S. cerevisiae hog1Δ null mutant (YSH444) was carried out with EhHOG. The salt tolerance of hog1Δ (YSH444 strain) is lower than that of the WT strain because GPD1 activity in response to osmotic stress is low when HOG1 (MAPK gene) is deleted (10). If EhHOG could complement the deletion of HOG1 in the S. cerevisiae hog1Δ strain, the osmotic tolerance of
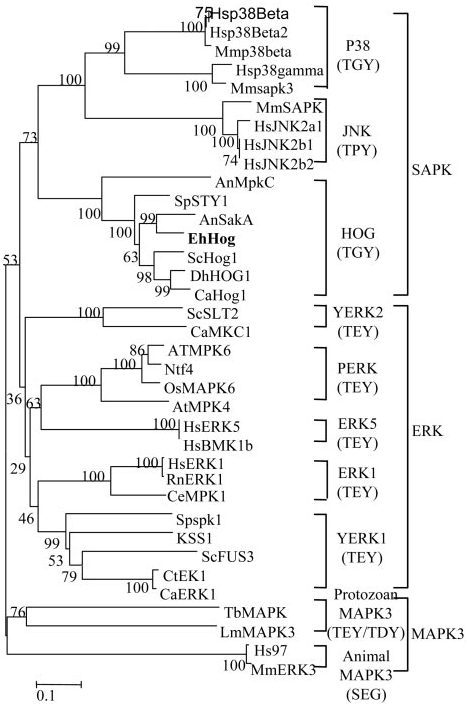
Fig. 2. Rooted phylogenetic tree of MAPK family constructed by the neigh- bor-joining method (19) based on amino acid sequences by the program MEGA 2.1 (www.megasoftware.net). The numbers at nodes are bootstrap confidence values based on 1,000 replicas. The tree is corrected with Poisson correction. Accession numbers of genes used are listed in Table 2, which is published as supporting information on the PNAS web site. P38, animal SAPK2 CLUSTER; JNK (c-jun N-terminal protein kinase), animal SAPK1 cluster; HOG, fungi SAPK cluster; PERK, plant ERK cluster; ERK1 and ERK5, human and animal ERK cluster; YERK1 and YERK2, yeast ERK1 and ERK2 clusters. The conserved dual phosphorylation motif TXY (threonine-variable amino acid-tyrosine) of each subfamily is shown in parentheses.
the transformant strain should be restored compared with the WT. The coding region of EhHOG was subcloned into a yeast 2µ vector pADNS and recombinant plasmid (pADNS/EhHOG) DNA was transformed into the hog1Δ (YSH444 yeast strain). The transformants were spotted on media containing 1 M NaCl, 1 M KCl, 2 M sorbitol, 0.4 M CaCl2, 20% Dead Sea water, and 300 mM LiCl (Fig. 3A). The mutant hog1Δ could not grow under these conditions, but the growth of the transformant strain (YSH444 strain containing EhHOG) was comparable with that of the WT strain under all these stress situations apart from 300 mM LiCl treatment where the transformant strain grew faster than the WT (Fig. 3). These results indicate that EhHOG has a similar function to the S. cerevisiae MAPK gene (HOG1) in respect to salt tolerance in general, whereas Li+ tolerance directed by EhHOG is clearly higher than that of HOG1 of the WT.
The S. cerevisiae hog1Δ null mutant has an aberrant cell
morphology under osmotic stress due to large multinucleated cells with multiple elongated buds (12). However, hog1Δ mutant containing EhHOG showed a normal cell shape (Fig. 4). This result indicated that the abnormal cell shape of hog1Δ mutants could be rescued by the expression of EhHOG.
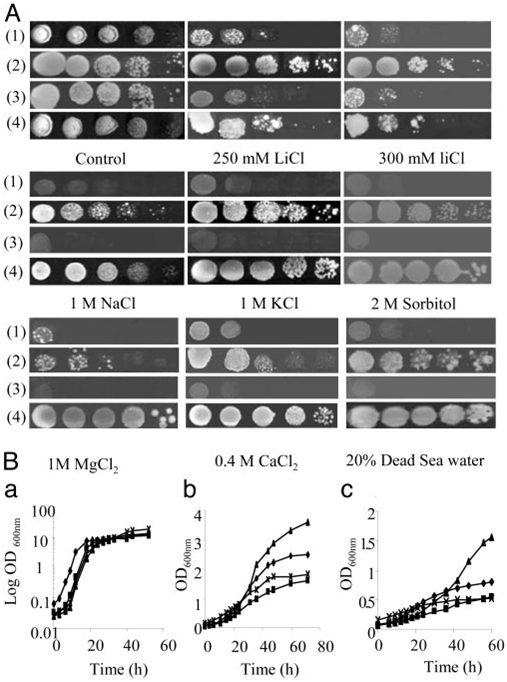
Fig. 3. Growth of yeast strains in YPD medium containing various salts. (A) Growth of S. cerevisiae hog1Δ (1), hog1Δ containing EhHOG (2), hog1Δ containing empty plasmid pADNS (3), and WT strains (4) in the presence of LiCl, NaCl, KCl, MgCl2, CaCl2, Dead Sea water, and sorbitol. Serial 10-dilutions of saturated cultures (OD600 = 2) were spotted onto YPD plates (control) sup- plemented with LiCl, NaCl, KCl, MgCl2, CaCl2, Dead Sea water, and sorbitol at the concentrations indicated. The plates then were incubated at 30°C. (B) Growth of yeast strains in YPD (a), 250 mM LiCl (b), and 300 mM LiCl (c). Diamonds, WT; squares, mutant (hog1Δ); triangles, hog1Δ containing EhHOG; crosses, hog1Δ containing empty vector. Growth of cells was estimated by measuring turbidity at 600 nm.
During salt stress, intracellular glycerol content in S. cerevisiae hog1Δ mutant increased to a much lower level than in the WT strain and the transformant (M/pEhHOG) (Fig. 5). Whereas compared with the WT, the transformant (M/pEhHOG)
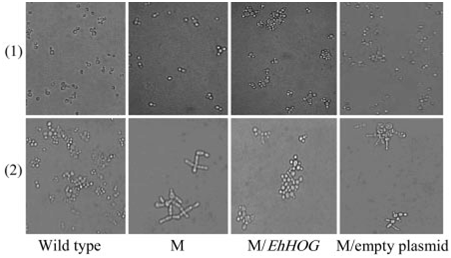
Fig. 4. Morphology of S. cerevisiae strains in the presence of 250 mM LiCl. WT strain (YSH6.142–3A), M (hog1Δ), M/EhHOG, and M/empty plasmid strains were grown in YPD (1) or with 250 mM LiCl (2).
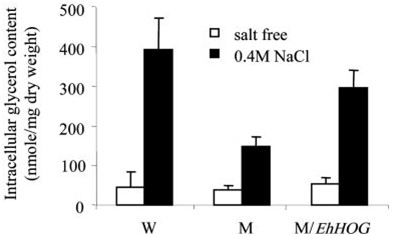
Fig. 5. Change of intracellular glycerol content in S. cerevisiae strains WT (W), hog1Δ mutant (M), and mutant containing EhHOG gene (M/pEhHOG). Cells were grown in SD media and harvested at OD600 0.5– 0.8. The cells were resuspended in fresh media with or without 0.4 M NaCl and incubated further for1h at 30°C, and then intracellular glycerol was measured. The results were obtained from three independent experiments with the same strains and the standard errors of means are indicated.
showed only a similar level of increase in intracellular glycerol content when it was stressed in 0.4 M NaCl (Fig. 5).
EhHOG cDNA complemented the hog1Δ null mutation and restored cell growth (Fig. 3A) and morphology (Fig. 4) under salt and osmotic stress conditions, demonstrating the function of MAPK (HOG pathway) in the osmosensing signal-transduction pathway.
The transformant appeared to have different growth rates on various salt media (Fig. 3), although it restored hog1Δ null mutant growth on diverse salt media. On 250- and 300-mM LiCl YPD plates, the transformant grew better than the WT. In 1 M MgCl2 and 0.4 M CaCl2 YPD media, the WT strain grew better than the transformant. In 1 M KCl YPD plate, the transformant grew better than in 1 M NaCl. It is possible that Na+ has more toxicity than K+. Uptake of K+ is beneficial for salt tolerance because K+ counteracts the inhibitory effects of Na+ on enzy- matic systems (30). As for the transformant, which has higher tolerance to LiCl, this feature is clearly derived from the specific sequence of EhHOG.
It is highly likely that there is an equivalent HOG pathway in
E. herbariorum cells. Moreover, this hog1Δ/EhHOG transfor- mant appears more tolerant than the WT strain under 250 and 300 mM LiCl (Fig. 3). The EhHOG transformant also increased intracellular glycerol content during salt stress (Fig. 5). However, EhHOG did not improve salt tolerance in WT S. cerevisiae (data not shown). These results suggest that EhHOG has similar function to HOG1 in S. cerevisiae to regulate the GPD1 gene, a second gene involved in glycerol biosynthesis in S. cerevisiae (31). Most microorganisms will not be able to survive in E. her- bariorum’s habitat, the Dead Sea, because of stress caused by unusually high salt concentration. E. herbariorum’s adaptive complex to this harsh environment could be governed by a range of alterations in the DNA level accumulated in the last 70,000 years. EhHOG’s contribution to this adaptive complex may not be obvious in our current approach because E. herbariorum’s higher salt tolerance could well result from a complicated network of many genes and pathways interacting with each other. Nevertheless, the higher Li+ tolerance conferred by EhHOG to hog1Δ, in contrast to that of the WT yeast HOG1, stands out in our current work. In addition to its therapeutic effects, Li+ is highly toxic to microorganisms and plants at much lower con- centrations compared with that of Na+ (32). Li+ has been widely used as an analog for Na+ in research on stress with yeast and plants because of its high sensitivity to all cells with inherent advantage that low concentrations can be used, avoiding screen- ing for osmotolerance (33). The Li+ concentration in the Dead Sea water is ≈100 times higher than that of the normal seas,
Table 1. Concentration of major cations and lithium in sea and Dead Sea water
Source Sodium Potassium Magnesium Calcium Lithium
Sea water* 10.56 0.38 1.27 0.40 0.17 × 10—3
Dead Sea† 40.1 7.7 44 17.2 18 × 10—3‡
All values are in g/liter.
*From ref. 34.
†From ref. 35.
‡From ref. 36.
whereas the differentials with Na+ and K+ are only ≈3 and 20 times, respectively (Table 1) (34 –36). Clearly, Li+ could have exerted much higher selection pressure on any organism living in the Dead Sea in contrast to other ions such as Na+, K+, Ca2+, and Mg2+. Hence, EhHOG is probably an ingredient of E. herbariorum’s adaptive complex to the Dead Sea water with exceptionally high Li+ content. Nevertheless, the Li+ concentration in the Dead Sea is much lower than those used in our study. The Li+ concentration may not have been the only factor shaping up the specific character of EhHOG. It is more likely that it was the cumulative effect of many salts in the Dead Sea exerting various amounts of stress to any organism living there, with Li+ as the representative for trace elements with high biological impact.
Resistance of EhHOG Against Generalized Stresses. In addition to having a role in osmoregulation, EhHOG could be involved in responses to generalized stresses, such as heat stress and oxidative stress, in S. cerevisiae (37), S. pombe (12), and Candida albicans (38). During heat stress, the growth of hog1Δ strain recovered more slowly than the WT strain (Fig. 6). This result is similar to that of Winkler et al. (37). The transformant (hog1Δ/EhHOG) recovered faster than the hog1Δ strain, similar to the WT strain (Fig. 6). During oxidative stress, hog1Δ strain was more sensitive than the WT strain to H2O2, whereas the trans- formant (hog1Δ/EhHOG) showed a similar survival rate to the WT strain (Fig. 7B). Our study indicated that EhHOG has similar function to HOG1 in S. cerevisiae to withstand oxidative stress.
During freeze-thaw stress, the transformant (hog1Δ/ EhHOG) survival rate was higher than the WT strain, especially after 10 cycles (Fig. 7A). WT strain survival rate decreased rapidly after 10 cycles and reached a similar level to the hog1Δ strain in 20 cycles (Fig. 7A). During freezing, cells can be injured by physical factors, such as intracellular ice crystal formation and cellular dehydration (39). Intracellular ice crystals are believed to rupture the plasma membrane, resulting in a release of cellular components into the environment (40). Glycerol is probably the most widely used cryoprotectant that protects the cells during freezing by minimizing the detrimental effects of increased solute concentration and ice crystal formation (41). Recently, Izawa et al. (42) reported that intracellular glycerol in
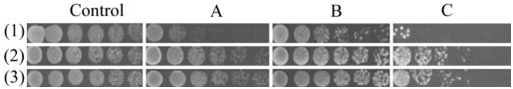
Fig. 6. Heat-stress tolerance of various yeast strains. Yeast cells M (mutant hog1Δ) (1), transformant M/EhHOG (2), and WT (3) were grown for 24 h in SD (minimal medium), and cell density was adjusted to OD600 at 1.8. Serial dilutions of 10 times were made. Ten microliters of each dilution was spotted on solid YPD plates (control). Cells on YPD plates were treated in 42°C for 1 day and allowed to recover at 28°C for 36 h (A) and 69 h (B); for 2-day heat stress, cells recovered at 28°C for 48 h (C).
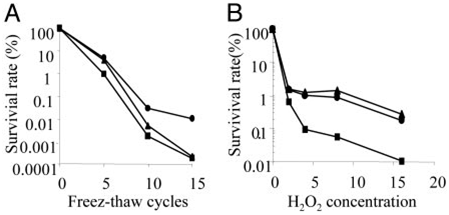
Fig. 7. Freeze- and oxidative-stress tolerance in various yeast strains. (A) Freeze-stress tolerance of cells was determined by measuring viability after the number of freeze-thaw cycles (—30°C and room temperature). (B) Cells were grown to OD600 1.0 –1.5 in SD medium at 30°C and then incubated for
1.5 h with increasing concentrations of H2O2 in SD. Percent survival was expressed relative to the initial viability before freezing or oxidative stress. Results are representative of three independent experiments. Filled triangles, WT; filled square, M, mutant hog1Δ; filled circle, M/EhHOG.
yeast plays an important role in tolerance to freezing-thawing stress. Despite this finding, because HOG1 regulates the expression of numerous genes, and glycerol is certainly not the only end product, the higher tolerances to Li+ and freezing-thawing brought by EhHOG may encompass contributions from other sources in addition to glycerol.
During the thawing process, cells suffer oxidative damage to cellular components by reactive oxygen species (43). Our results of the freeze-thaw stress were different from those of oxidative stress. This finding implies that a higher survival rate in the transformant (hog1Δ/EhHOG) than in the WT strain could not be due to high resistance to oxidative stress during freeze-thaw stress. Yeast cells may cope with freeze-thaw stress by the synthesis of stress protein (44) or metabolites such as trehalose and glycerol (45, 46). Trehalose stabilizes the intracellular water structure and cell membranes under stress conditions (45). Freeze–thaw-tolerant yeast strains had higher levels of trehalose (47), indicating that trehalose is a possible protectant in freeze-thaw stress. Hence, EhHOG in the transformant possibly pro- moted the expression of genes involved in glycerol and trehalose synthesis, which produced higher levels of the metabolites trehalose and glycerol synthesis in stress survival. This hypothesis could explain the results we obtained from studying freeze-thaw stress.
To further eliminate the possibility that the complementation is derived from the copy number of EhHOG or the ADH promoter in the construct (pADNS/EhHOG), HOG1 gene from WT yeast was isolated, and the construct (pADNS/ScHOG1) was made similarly to that of EhHOG (see Materials and Methods). As with EhHOG, S. cerevisiae hog1Δ was transformed with pADNS/ScHOG1, and the transformant was tested against all of the stresses as described above. The results showed that hog1Δ mutant transformed with EhHOG was superior to that of the hog1Δ mutant transformed with WT HOG1 in Li+ and freezing–thawing tolerances but not in other stresses (see Fig. 10, which is published as supporting information on the PNAS web site). The Dead Sea is already one of the most saline lakes on earth with total salt concentration increasing, while its sea level falls (48). Still, the fungi in the Dead Sea live in the gradually increasing saline environment. The content of most major elements is close to or already reaches saturation in the Dead Sea water. Trace elements, such as Li+, could play a more significant role in shaping up the genetic future of a few Dead Sea organisms as it may have done so in the past. The evolution of the Dead Sea from a freshwater Lisan lake to a lacustrine and hypersaline close lake (49) led to increasing adaptive salt resistance in the few
organisms that could survive in the Dead Sea, such as Archea (50), Dunaliella (2), and filamentous fungi (2).
Our results demonstrate that EhHOG could perform similarly to other HOGs under a range of stress environments. More importantly, yeast hog1Δ mutant, complemented with EhHOG, displayed higher tolerance to some types of salts and freezing as well. Evidently, we can attribute the superior tolerance to specific sequences of EhHOG in contrast to other HOGs. These specific sequences are most likely the products of strong selection pressure from the hypersalinity progressively increasing in the Dead Sea for the last 70,000 years of the Lisan lake duration, particularly during the last 15,000 years.
In the present study, EhHOG functioned in a distantly related yeast species without any obvious shortcomings. It is interesting that despite considerable differences between yeast and E. herbarium, we could achieve the functional expression of EhHOG, a MAPK homolog playing a central role in HOG signal transduction, in the yeast S. cerevisiae. Because Hog is well conserved from yeast to animals, there might be ample opportunity to introduce EhHOG into other eukaryotic genomes to improve stress tolerance. Moreover, other genes from E. her-variorum, as well as other organisms residing in the same environment, could prove useful for further genetic engineering efforts. It has been speculated that there is no HOG lineage in plants, because no plant HOG homolog has been found yet (29). Nevertheless, the MAPK homolog from Pisum sativum could complement the HOG mutant (51). It will be of great interest to see the interaction of EhHOG with a plant genome if stable integration can be achieved and thereby reinforce the exploitation of genetic resources of the Dead Sea fungi for crop improvement. Our preliminary results suggest that EhHOG might possess the potential to play a similar role, as shown here with a yeast hog1 mutant, in conferring better stress tolerance in transgenic Arabidopsis (data not shown).
Prospects. In conclusion, isolation and characterization of EhHOG show insight into the evolutionary strategies and genetic mechanism of high halotolerant in the Dead Sea fungus E. herbariorum. EhHOG also can enhance the resistance against multiple stress tolerances (salt, H2O2, low and high temperature), which constitutes a clear advantage in agriculture. These results undoubtedly open theoretical routes for the study of Dead Sea biodiversity of filamentous fungi and other organisms (e.g., Archea, the alga Dunaliella) in terms of stress genomics, proteomics, and phenomics. In addition, genetic resources in the Dead Sea also could be potentially harnessed for engineering transgenic organisms with enhanced stress tolerance, thereby advancing saline agriculture. This application is desperately relevant in view of the increasing desertification and salinization of our planet and the need for crop improvement and higher food production to overcome world hunger. We thank Prof. L. Adler (Go¨teborg University, Go¨teborg, Sweden) for contributing yeast strains and for help with glycerol measurement; Dr. Y. Kassir (Institute of Technion, Haifa, Israel) for contributing the vector; and Profs. M. Gustin, M. Montero Lomeli, and B. Horwitz for commenting on the manuscript and leading to its improvement. This work was supported in part by the Authority of Graduate Studies of the University of Haifa, the Israel Discount Bank Chair of Evolutionary Biology, and the Ancell Teicher Research Foundation for Molecular Genetics and Evolution.
- Nevo, E., Oren, A. & Wasser, S. P. (2003) Fungal Life in the Dead Sea (Gantner, Ruggell, Germany).
- Kis-Papo, T., Grishkan, I., Oren, A., Wasser, S. P. & Nevo, E. (2001) Mycol. Res. 105, 749 –756.
- Yancey, P. H., Clark, M. K., Hand, S. C., Bowlus, R. D. & Somero, G. N. (1982) Science 217, 1214 –1222.
- Mager, H. G. & Varela, J. C. S. (1993) Mol. Microbiol. 10, 253–258. 6. Seger, R. & Krebs, E. G. (1995) FASEB J. 9, 726 –735.
- Banuett, F. (1998) Microbiol. Mol. Biol. Rev. 62, 249 –274.
- Gustin, M. C., Albertyn, J., Alexander, M. & Davenport, K. (1998) Mol. Biol. Rev. 62, 1264 –1300.
- Brewster, J. L., de Valoir, T., Dwyer, N. D., Winter, E. & Gustin, M. C. (1993)
- Science 259, 1760 –1763.
- Albertyn, J., Hohmann, S., Thevelein, J. M. & Prior, B. A. (1994) Mol. Cell. Biol. 14, 4135– 4144.
11. Norbeck, J. & Blomberg, A. (1997) J. Biol. Chem. 272, 5544 –5554.
- Degols, G., Shiozaki, K. & Russell, P. (1996) Mol. Cell. Biol. 16, 2870 –2877.
- Zhang, Y., Lamm, R., Pillonel, C., Lam, S. & Xu, J. R. (2002) Appl. Environ. Microbiol. 68, 532–538.
- Han, J., Lee, J. D., Bibbs, L.& Ulevitch, R. J. (1994) Science 265, 808 – 811.
- Sherman, F. (1991) Methods Enzymol. 194, 3–21.
- Mcalpin, C. E. & Mannarelli, B. (1995) Appl. Environ. Microbiol. 61, 1068 – 1072.
- Philippsen, P., Stotz, A. & Scherf, C. (1991) Methods Enzymol. 194, 170 –172.
- Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Woodbury, NY), 2nd Ed.
- 19. Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406 – 425.
- Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17,1244 –1245.
- Gietz, R. D. & Woods, R. A. (2002) Methods Enzymol. 350, 87–92.
- Andr´e, L., Nilsson, A. & Adler, L. (1998) J. Gen. Microbiol. 144, 669 – 677.
- Arnold, C. & Hodgson, I. J. (1991) PCR Methods Applications 1, 39 – 42.
- Cano, E. & Mahadevan L. C. (1995) Trends Biochem. Sci. 20, 117–122.
- Kumar, S., McLaughlin, M. M., McDonnell, P. C., Lee, J. C., Livi, G. P. & Yong P. R. (1995) J. Biol. Chem. 270, 29043–29046.
- Boquslawski, G. (1992) J. Gen. Microbiol. 138, 2425–2432.
- Tanoue, T., Adachi, M., Moriguchi, T. & Nishida, E. (2000) Nat. Cell. Biol. 2,110 –116.
- Enslen, H. & Davis, R. J. (2001) Biol. Cell 93, 5–14. 29. Ku¨ltz, D. (1998) J. Mol. Evol. 46, 571–588.30. Serrano, R. (1996) Int. Rev. Cytol. 165, 1–52.
- Erikksson, P., Andre, L., Ansell, R., Blomberg, A. & Adler, L. (1995) Mol. Microbiol. 17, 95–107.
- Masuda, C. A., Xavier, M. A., Mattos, K. A., Galina, A. & Montero-Lomeli, M. (2001) J. Biol. Chem. 276, 37794 –37801.
- Bohnert, H. J., Nelson, D. E. & Jensenayb, R. G. (1995) Plant Cell 7, 1099 –1111.
- Mason, B. (1974) Principles of Geochemistry (Wiley, New York), 3rd Ed., p. 329.
- Beyth, M. (1980) in Hypersaline Bringes and Evaporitic Environments, ed. Nissenbaum, A. (Elsevier, Amsterdam), pp. 155–165.36. Nissenbaum, A. (1977) Chem. Geol. 19, 99 –111.
- Winkler, A., Arkind, C., Mattison, C. R., Burkholder, A., Knoche, K. & Ota,
- (2002) Eukaryotic Cell 1, 163–173.
- Alonso-Monge, R., Navarro-Garcia, F., Roman, E., Negredo, A. E., Eisman, B., Nombla, C. & Pla, J. (2003) Eukaryotic Cell 2, 351–361.
- Tanghe, A., Dijck, V. P. & Thevelein, J. M. (2003) Appl. Microbiol. 53, 129 –161. 40. Mazur, P. (1977) Cryobiology 14, 251–272.
- Lovelock, J. E. (1953) Biochim. Biophys. Acta 11, 28 –36.
- Izawa, S., Sato, M., Yokoigawa, K. & Inoue, Y. (2004) Appl. Microbiol. Biotechnol. 66, 108 –114.
- Park, J. I., Grant, C. M., Davies, M. J. & Dawes, I. W. (1998) J. Biol. Chem.36, 22921–22928.
- Komatsu, Y., Kaul, S. C., Iwahashi, H. & Obuchi, K. (1990) FEMS Microbiol. Lett. 72, 159 –162.
- Iwahashi, H., Obuchi, K., Fujii, S. & Komatsu, Y. (1995) Cell. Mol. Biol. 41,763–769.
- Lewis, J. G., Learmonth, R. P. & Watson, K. (1995) Microbiol. 141, 687– 694.
- Hino, A., Mihara, K., Nakashima, K. & Komatsu, Y. (1990) Appl. Environ. Microbiol. 56, 1386 –1391.
- Oren, A. & Gavrieli, I. (2002) SIL News 35, 1–5.
- Bartov, Y. (2003) in Fungal Life in the Dead Sea, eds. Nevo, E., Oren, A. & Wasser, S. (Gantner, Ruggell, Germany), pp. 26 – 44.
- Oren, A. (2003) in Fungal Life in the Dead Sea, eds. Nevo, E., Oren, A. & Wasser, S. (Gantner, Ruggell), pp. 117–140.
- Popping, B., Gibbons, T. & Watson, M. D. (1996) Plant Mol. Biol. 31,355–363.







